HTRC Workset Toolkit
Here you can read up on all the functionalities the Workset Toolkit affords data capsule researchers.
HTRC Workset Toolkit is a command line interface for use in the HTRC Data Capsule environment. It streamlines access to the /wiki/spaces/COM/pages/43286551 and includes utilities to pull OCR text data and volume metadata into a capsule. Additionally, it allows a researcher to point OCR text data to analysis tools that are also available in the capsule.
Capsules created after March 18, 2018 contain the Toolkit by default. If you created your capsule before March 18, 2018, please skip to Getting the HTRC Workset Toolkit below.
Additional documentation is also available here: https://htrc.github.io/HTRC-WorksetToolkit/cli.html
Usage
The HTRC Workset Toolkit has four primary functions which allow users to download metadata and OCR data, run analysis tools, and export lists of volume IDs from the capsule.
- Volume Download
htrc download
- Metadata Download
htrc metadata
- Pre-built Analysis Workflows
htrc run
- Export of volume lists
htrc export
Identifying volumes
Each command expects a workset path, which is how you point to the volume(s) you would like to analyze. The Toolkit accepts several different forms of identifiers for the workset path, as described in the following table. You can choose to use whichever is the most conducive to your research workflow. You don't need to specific which kind of workset path you will be using, you can simply include the identifier (e.g. the HathiTrust ID or the HathiTrust Catalog URL) in your command.
| Workset path | Example | Note |
|---|---|---|
| File of volume IDs | /home/dcuser/Downloads/collections.txt | |
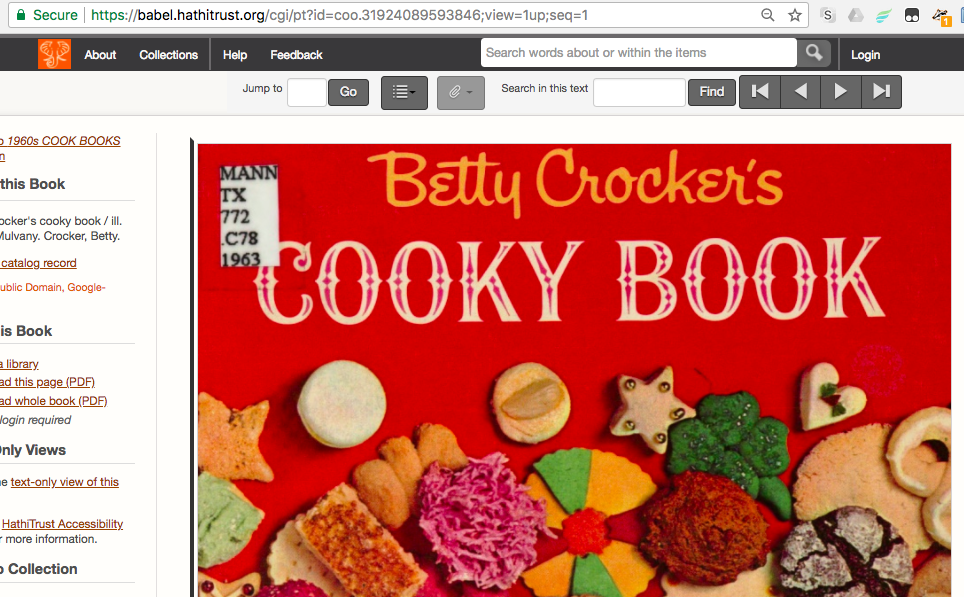
| HathiTrust ID | mdp.39015078560078 | |
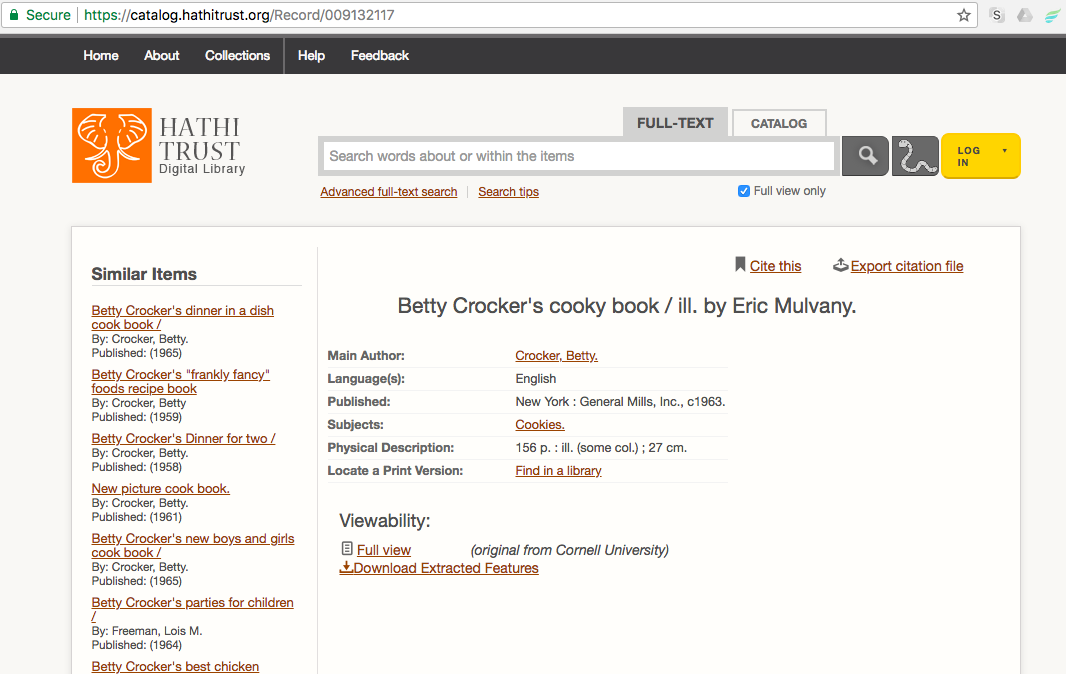
| HathiTrust Catalog ID | 001423370 | |
| HathiTrust URL | https://babel.hathitrust.org/cgi/pt?id=mdp.39015078560078;view=1up;seq=13 | must be in quotes in command |
| Handle.org Volume URL | https://hdl.handle.net/2027/mdp.39015078560078 | must be in quotes in command |
| HathiTrust Catalog URL | https://catalog.hathitrust.org/Record/001423370 | must be in quotes in command |
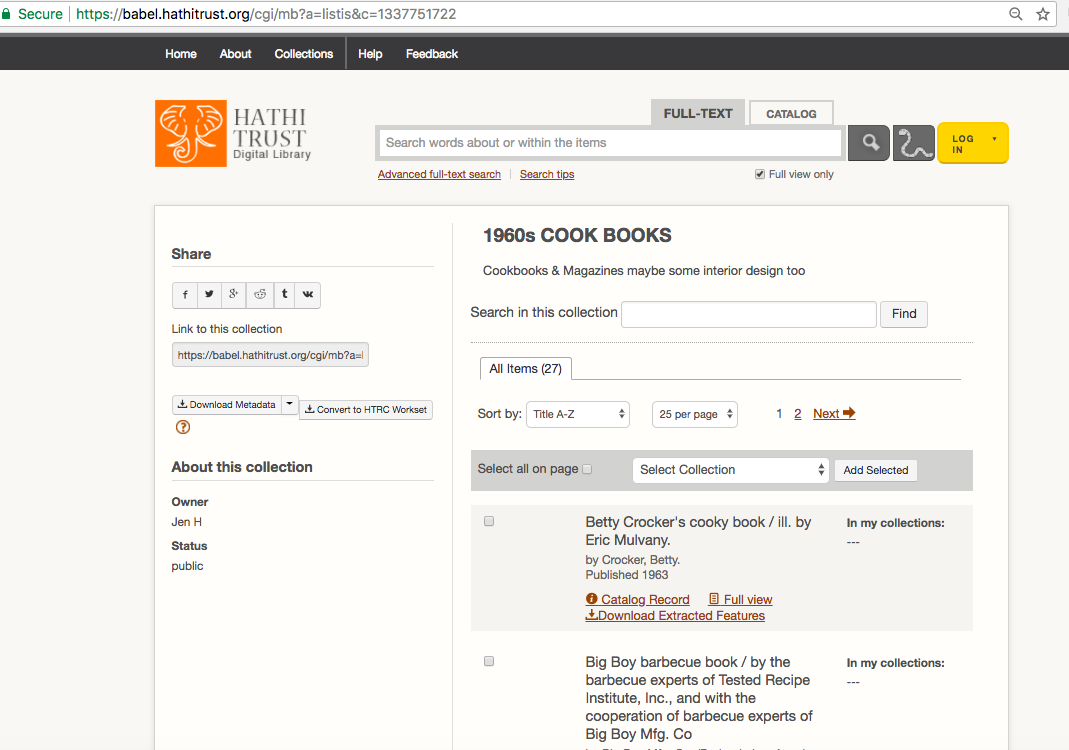
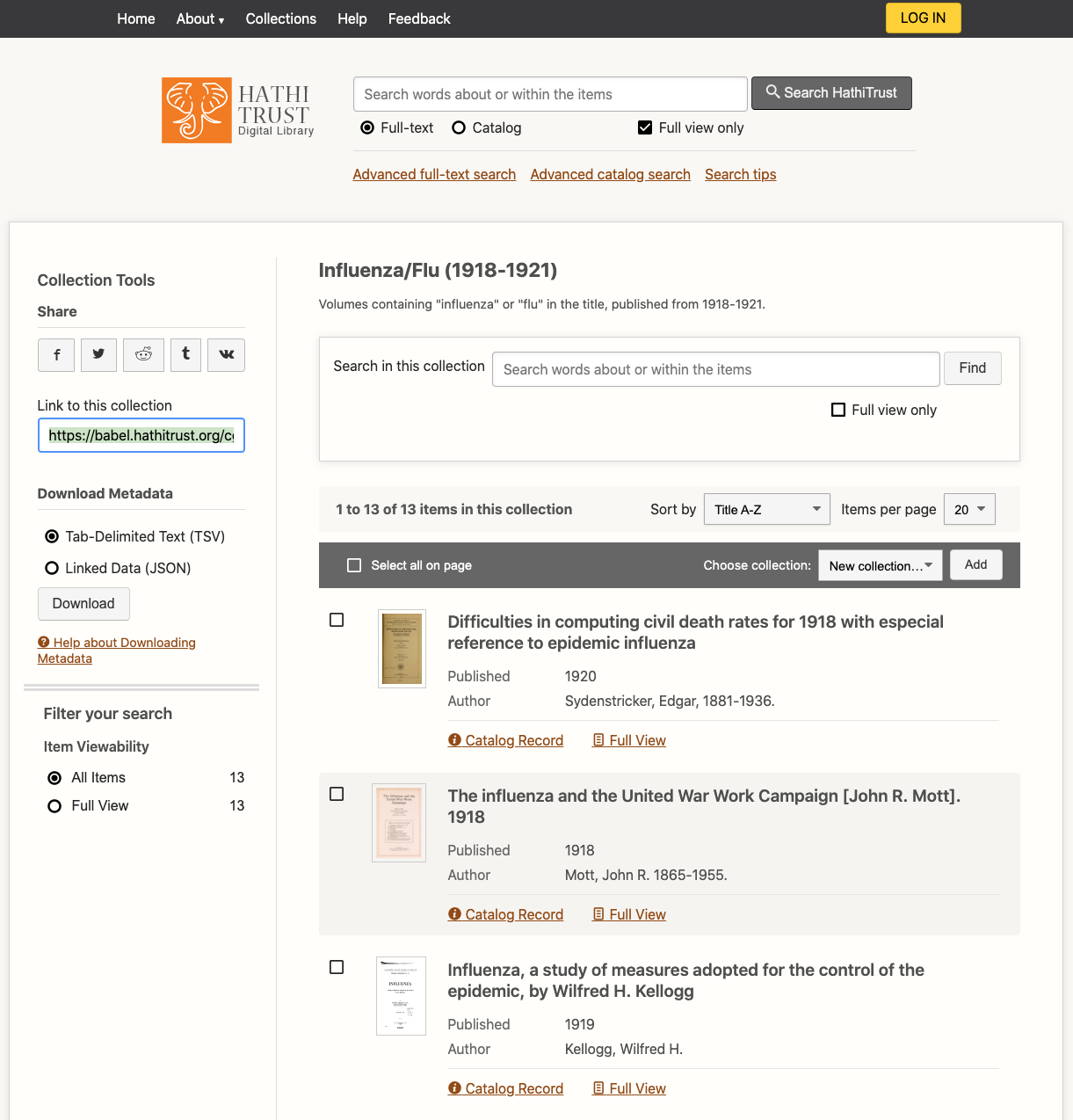
| HathiTrust Collection Builder URL (for public collections only) | https://babel.hathitrust.org/shcgi/mb?a=listis;c=696632727 | must be in quotes in command |
Building a dataset
To customize your dataset, you will need to search and build a collection in HathiTrust. Or use other metadata sources, including HathiFiles, to create a list of HathiTrust volume IDs. HathiTrust IDs are the unique identifier for each volume, and you can see an example in the table above.
If you are importing data using a file of volume IDs, your file should in txt format with one ID per line.
You can access data that corresponds to the permissions level associated with your capsule. Only capsules that have been granted full corpus access are able to import any volume from the HathiTrust collection. Demo and general research capsules can only import public domain (i.e. "Full View") items.
Volume Download
The basic form to import OCR data is to use the "htrc download" function. When you run the command, you have your choice of several arguments that impact how the data is transferred and you also provide the workset path. This command will only work in secure mode; it won't work in maintenance mode for security reasons.
The format for the command looks like this:
htrc download [-f] [-o OUTPUT] [-c] [WORKSET PATH]
The brackets indicate optional text and/or text that should be changed before you run the command. The named arguments, which are the "flagged" letters in the command, are described in the following table:
| Named argument | What it does | What happens if it's NOT included |
| -f, --force | Remove folders if they exist | Each volume in your dataset will be located in it's own folder |
| -o, --output | Indicates that you will be choosing a directory location where the files should go. Should be followed by the in-capsule directory path of your choice. You can call the destination folder anything you like. | Data will be sent to “/media/secure_volume/workset/” |
| -c, --concat | Concatenate a volume’s pages in to a single file. | Page files will not be concatenated |
| -pg, --pages | Download only certain pages from a volume. | All page files for each volume will be downloaded |
| -m, --mets | Includes the METS file for each volume in your download request. Cannot be used with both -pg (pages) and -c (concatenate). | METS files are not downloaded |
| -h, --help | Displays the help manual for the toolkit. | Help manual is not displayed |
| -hf, --headfoot | Removes headers and footers from pages of downloaded volumes. | All volume pages will be downloaded with headers and footers still included |
| -hfc, --headfootcon | Removes headers and footers from pages of downloaded volumes then concatenates the pages into a single txt file. | All volume pages will be downloaded with headers and footers included and each page will be an individual file |
| -s , --skip-removed-hf | This will skip the creation of a .csv file that contains the removed headers/footers for each volume when used in conjunction with the -hf or -hfc flags. | If not included in conjunction with the -hf or -hfc flags the .csv files will be created and saved to the output folder by default. If used without either the -hf or -hfc flags it will produce an error. |
| -w, --window-size | This flag, followed by an integer, (-w 5) and used in conjunction with the -hf or -hfc flags indicates how many pages ahead does the header/footer extractor algorithm look to find potential matching headers/footers. The default is 6 pages. A higher value gives potentially more accurate results on lower quality OCR volumes at the expense of runtime. | If not included in conjunction with the -hf or -hfc flags the page window will default to 6 pages. |
| -msr, --min-similarity-ratio | When used in conjunction with the -hf or -hfc flags the -msr flag followed by a float between 0 and 1 ( -msr .9) will change the minimum string similarity ratio required for the Levenshtein distance fuzzy-matching algorithm to declare that two headers are considered 'the same'. The higher the value, up to a max of 1.0, the more strict the matching has to be; lower values allow for more fuzziness to account for OCR errors. | If not included in conjunction with the -hf or -hfc flags the minimum similarity rate defaults to .7 (70%). |
| --parallelism | This flag followed by an integer (with the maximum being the number of vcpus in the data capsule, for a research capsule this is 4, but can be less, and the maximum for a demo capsule is 2 and, used in conjunction with the -hf or -hfc flags, changes the max number of concurrent tasks to start when downloading or removing headers/footers. | If not included in conjunction with the -hf or -hfc flags the default is to use all of the available cores of the system. |
| --batch-size | Changes the max number of volumes to download at a time from DataAPI when followed by an integer. | Defaults to 250 volumes at a time. |
Examples
Be sure that you are in secure mode before trying these!
Getting the HTRC Workset Toolkit
- Capsules created prior to March 18, 2018 did not contain the HTRC Workset Toolkit command line interface by default, and user-installed versions of the Toolkit may now be out of date and fail to run. To ensure you are running the most up-to-date version of the Toolkit, please follow these steps to uninstall the current version and re-install the latest version:
Check whether you have the correct Python version installed in your capsule by typing the command indicated below on the command line in your capsule. The HTRC Workset Toolkit requires the Anaconda Python distribution, which is likewise standard in all recently-created capsules. And while the Toolkit is compatible with both Python 2.7 and 3.6, we recommend using the 3.6 version for future compatibility.
Check python versiondcuser@dc-vm:~$ python --version
You should see thisPython 3.6.0 :: Anaconda 4.3.1 (64-bit)
Some users may have self-installed the Toolkit in their capsules prior to March 18, 2018. If you have already installed the HTRC Workset Toolkit, uninstall it by using pip, as indicated below.
Uninstall the toolkitdcuser@dc-vm:~$ pip uninstall htrc
Install the latest version of the HTRC Workset Toolkit.
Install the toolkitdcuser@dc-vm:~$ pip install htrc
Please note that updating may affect the versions of packages listed at HTRC-WorksetToolkit/setup.py that you are running in your capsule, and therefore should be done with care for your existing workflows. Contact htrc-help@hathitrust.org for assistance upgrading your version of the Toolkit.